Tom van den Bergh understands proteins. The 3DM system he co-developed accurately charts how proteins work and how you can manipulate them
Life on earth is impossible without proteins, which play a key role in all processes in living cells. So they are present in staggering numbers and great diversity. And due to mutations, no two proteins are exactly the same. A human being couldn’t possibly keep track of them, but a computer can. Tom van den Bergh recently obtained his doctorate in this field.
The focus of his study is the 3DM programme, which collects, organizes and analyses protein data. He worked on it a lot during his Bioinformatics studies in Leiden, where he did both his Bachelor’s and his Master’s theses on research that resulted in 3DM, the life’s work of his current boss Henk-Jan Joosten. Van den Bergh initially worked in the basement of the Microbiology lab on De Dreijen campus, where Joosten started a small company called Bio-Prodict in 2008.
‘I did my MSc there, and then went on to work for the company,’ says Van den Bergh. But the basement soon became too small, and the company moved to Nijmegen, where it has been for over 10 years. ‘I did my PhD part-time while I was working for Bio-Prodict. That’s why it took a while. I think it took me about eight years, altogether.’ He had finished his thesis two years ago, actually. But his defence was postponed due to Covid-19.
Folded
‘3DM is the 3D version of a system that brings together all the available information about protein families,’ Van den Bergh explains. ‘And a protein family consists of all the proteins that are folded in the same way in 3D. A protein is a chain of amino acids that folds itself into an effective 3D structure in response to various physical and chemical forces. The folding creates cavities, cracks and places where reactions can occur.’
But proteins mutate in the course of evolution. ‘So there are very many proteins that differ slightly in their amino acid sequence, but still fold in the same way,’ Van den Bergh continues. ‘There is a lot of data on amino acid sequences online. With 3DM we superimpose the structures of those comparable proteins over each other. This alignment of 3D structures happens quite literally on the computer screen. This kind of alignment can involve hundreds of thousands of proteins. The images show which parts of the protein, and therefore which amino acid sequences, are essential for the functioning of the molecule.
‘If you have a protein no one knows much about, you can skip a whole lot of research thanks to this system. Based on its structure and similarity to known proteins, you can deduce all kinds of information about the unknown protein,’ explains Van den Bergh. ‘Just by comparing the amino acid sequence, you gain a lot of information. Positions in the protein that always remain the same are apparently important for the functioning of the protein. These positions are of course also subject to mutations, but such mutations disappear due to evolutionary pressure, because the organism does not survive.’
Predictions
There are very many applications of 3DM. Biotechnologists, for example, make eager use of it to develop new or improved enzymes. Van den Bergh: ‘Once you have mapped all those mutations, you can ask the simple question: which one has an effect on the specificity of the protein? Where do you have to change the enzyme to make it also work with a slightly different substance? Or to make it work faster or at a higher temperature, or remain more stable? Two weeks of work on a 3DM system like this can save you six months’ work in the lab’.
Twee weken werk aan zo’n 3DM-systTwo weeks of work on a 3DM system like this can save you six months’ work in the lab
The medical applications are at least as interesting. Using machine learning, Van den Bergh developed a programme that predicts the likelihood of a mutation at a particular location in the human genome leading to disease. ‘If you can predict which mutation will change a protein in a desired direction, you can also predict which mutation will negate the effect of a protein. I have done this for three proteins that are involved in LQT syndrome, a condition that can lead to cardiac arrhythmia and an increased risk of cardiac arrest. An abnormal protein does not necessarily mean that you have LQT. Each genome is slightly different, due to natural variation. No doctor wants to treat someone who is actually healthy. The predictor tells you whether a change in the protein is likely to be pathogenic or not.’
Exome
That was the state of affairs two years ago. Van den Bergh: ‘We have now made this predictor for the entire human exome, in other words for all human proteins. We can predict the effect of every mutation in a protein.’ And that was no small task. Humans have 20,000 genes that code for proteins. One gene can also code for several protein variants, which can be traced back to almost 6000 protein families.
It sounds like science fiction: predicting the risk of disease on the basis of your genome. ‘But a lot of questions remain unanswered,’ says Van den Bergh reassuringly. ‘Our predictor only says whether a mutation could lead to illness. It cannot tell you how ill you will become or exactly which disease you will get. But things are certainly developing extremely fast.’

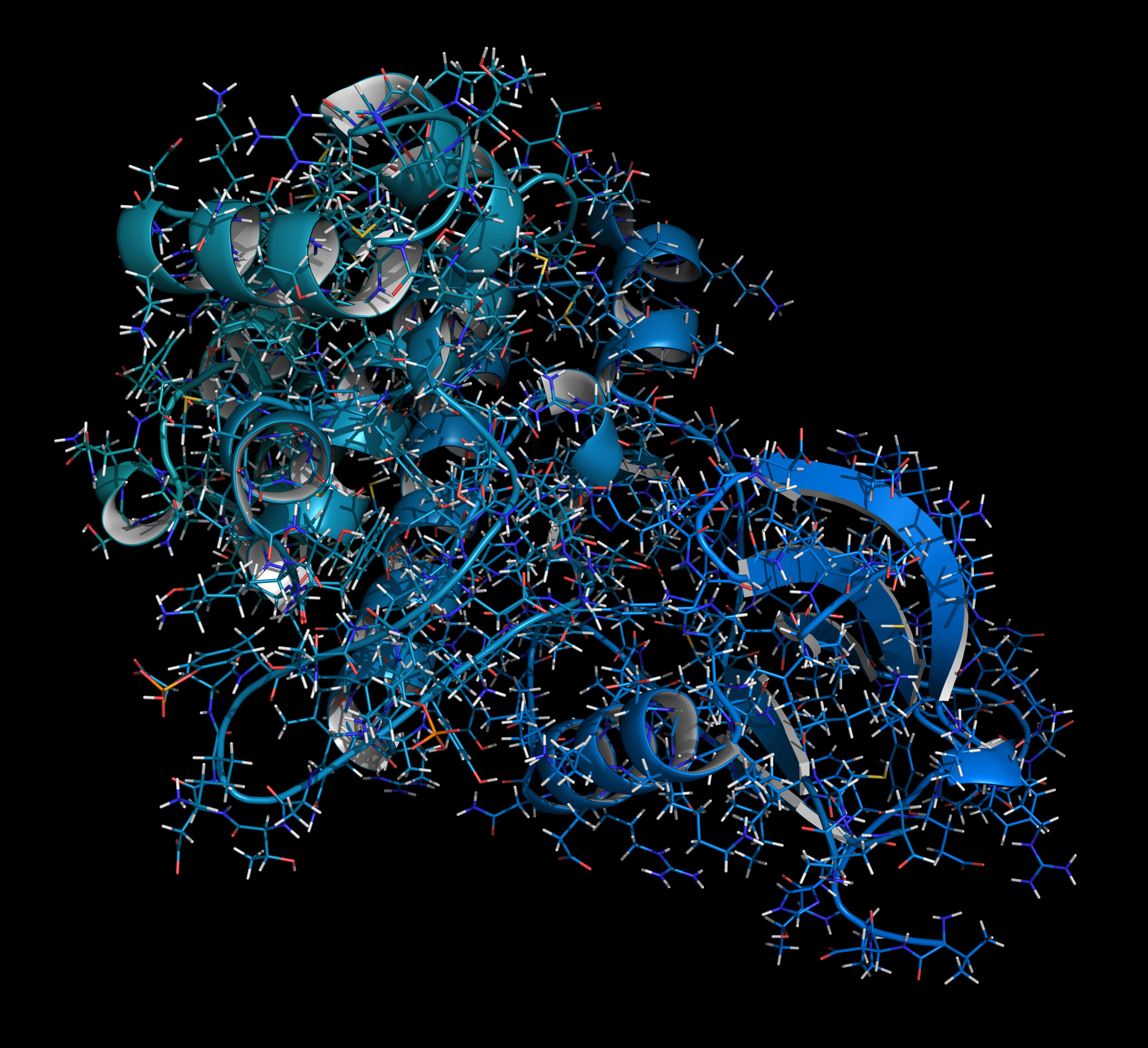 Photo: Shutterstock
Photo: Shutterstock